# Load libraries
library(readr)
library(tidyr)
suppressPackageStartupMessages(library(dplyr))
library(ggplot2)
library(linearpackcircles)
# Data
DF = read_csv("https://raw.githubusercontent.com/owid/covid-19-data/master/public/data/owid-covid-data.csv", show_col_types = FALSE) %>%
drop_na(total_cases_per_million, total_deaths_per_million) %>%
filter(date == max(date)) # Keep only most recent dataSimple visualization
# Create Plot
plot1 = linearpackcircles(DF,
ID_var = "location",
group_var = "continent",
area_var = "total_cases_per_million",
x_var = "total_deaths_per_million",
separation_factor = 200, # Separation between groups (i.e. continents)
width_plot = 2000, # width "units"
height_group = 100, # height within each group
label_circles = TRUE,
max_overlaps = 8,
size_text = 2,
area_multiplier = 1000)
# Add title, caption...
plot1 +
labs(title = "COVID deaths per million",
x = "Deaths per million",
caption = "Diameter is cases per million \n Data from https://github.com/owid/covid-19-data \nBy @gorkang")
#> Warning: ggrepel: 48 unlabeled data points (too many overlaps). Consider
#> increasing max.overlaps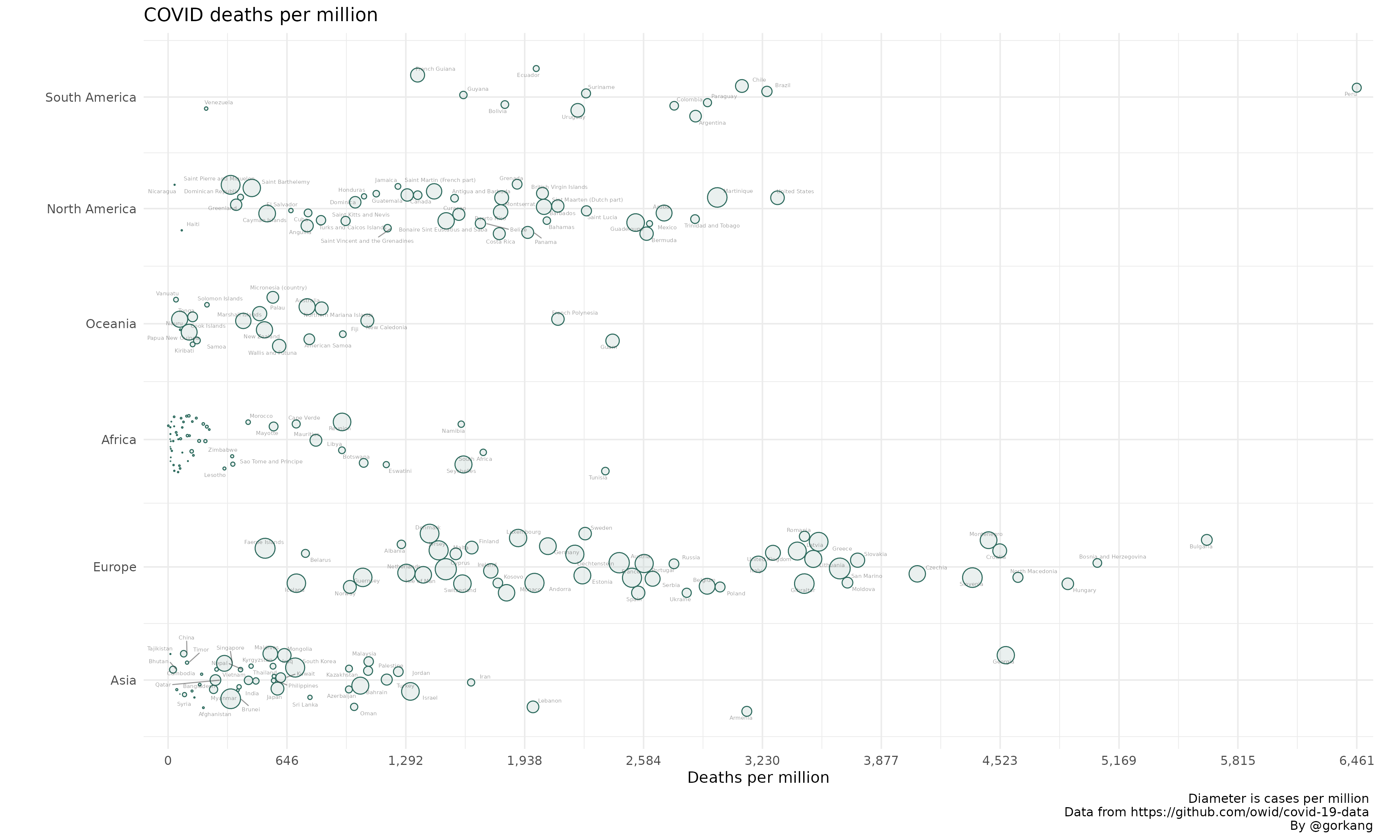
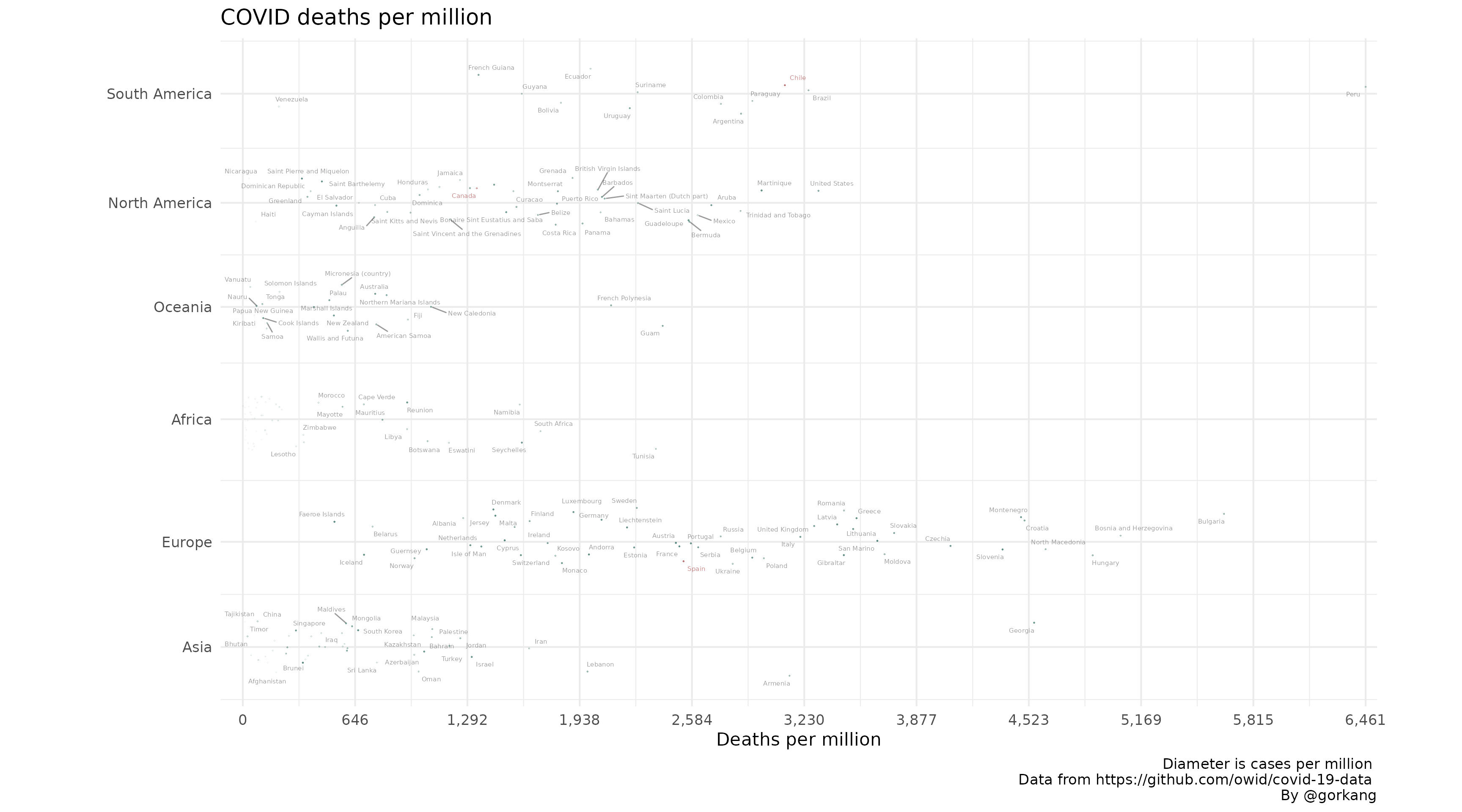
Highlight
You can highlight any number of elements (i.e. counties).
# Create Plot
plot1 = linearpackcircles(DF,
ID_var = "location",
group_var = "continent",
area_var = "total_cases_per_million",
x_var = "total_deaths_per_million",
separation_factor = 200,
width_plot = 2000,
height_group = 100,
label_circles = TRUE,
max_overlaps = 8,
size_text = 2,
highlight_ID = c("Canada", "Chile", "Spain"))
# Add title, caption...
plot1 +
labs(title = "COVID deaths per million",
x = "Deaths per million",
caption = "Diameter is cases per million \n Data from https://github.com/owid/covid-19-data \nBy @gorkang")
#> Warning: ggrepel: 67 unlabeled data points (too many overlaps). Consider
#> increasing max.overlaps