13 Experimental design
13.1 Tamaño de muestra y efecto detectado
En esta simulación se muestra como con tamaños de muestra pequeños, se sobreestima el tamaño de efecto real. Como se puede ver en el codigo, solo se muestran los estudios simulados donde la correlacion es significativa. Ver el blog de Jim Grange para una explicación más detallada.
# https://jimgrange.wordpress.com/2017/03/06/low-power-effect-sizes/
#------------------------------------------------------------------------------
rm(list = ls())
set.seed(50)
# function for generating random draws from multivariate distribution
# n = number of draws; p = number of variables
# u = mean of each variable; s = SD of each variable
# corMat = correlation matrix
mvrnorm <- function(n, p, u, s, corMat) {
Z <- matrix(rnorm(n * p), p, n)
t(u + s * t(chol(corMat)) %*% Z)
}
#------------------------------------------------------------------------------
#------------------------------------------------------------------------------
### simulation setup
# declare simulation parameters
means <- c(100, 600)
sds <- c(20, 80)
n_sims = 1000
# declare correlation coefficient & generate cor. matrix
cor <- 0.3
cor_mat <- matrix(c(1, cor,
cor, 1), nrow = 2, ncol = 2, byrow = TRUE)
# sample sizes to simulate
sample_sizes <- c(10, 20, 30, 50, 85, 170, 500, 1000)
# number of simulations
# create variable to store data in
final_data <- matrix(nrow = n_sims, ncol = length(sample_sizes))
colnames(final_data) <- sample_sizes
#------------------------------------------------------------------------------
#------------------------------------------------------------------------------
### simulation execution
for(i in 1:length(sample_sizes)){
for(j in 1:n_sims){
# get the experiment data
sim_data <- mvrnorm(sample_sizes[i], p = 2, u = means, s = sds,
corMat = cor_mat)
# perform the correlation
sim_cor <- cor.test(sim_data[, 1], sim_data[, 2], method = "pearson")
# if the correlation is significant, store the effect size
if(sim_cor$p.value < 0.05){
final_data[j, i] <- as.numeric(round(abs(sim_cor$estimate), 3))
}
}
}
#------------------------------------------------------------------------------
#------------------------------------------------------------------------------
## draw the plot
boxplot(final_data, na.action=na.omit, xlab = "Simulated Sample Size",
ylab = "Measured Effect Size")
abline(h = cor, col = "red", lty = 2, lwd = 3)
legend("topright", "True Effect Size", cex = 1.5, lty = 2, bty="n",
col = "red", lwd = 3)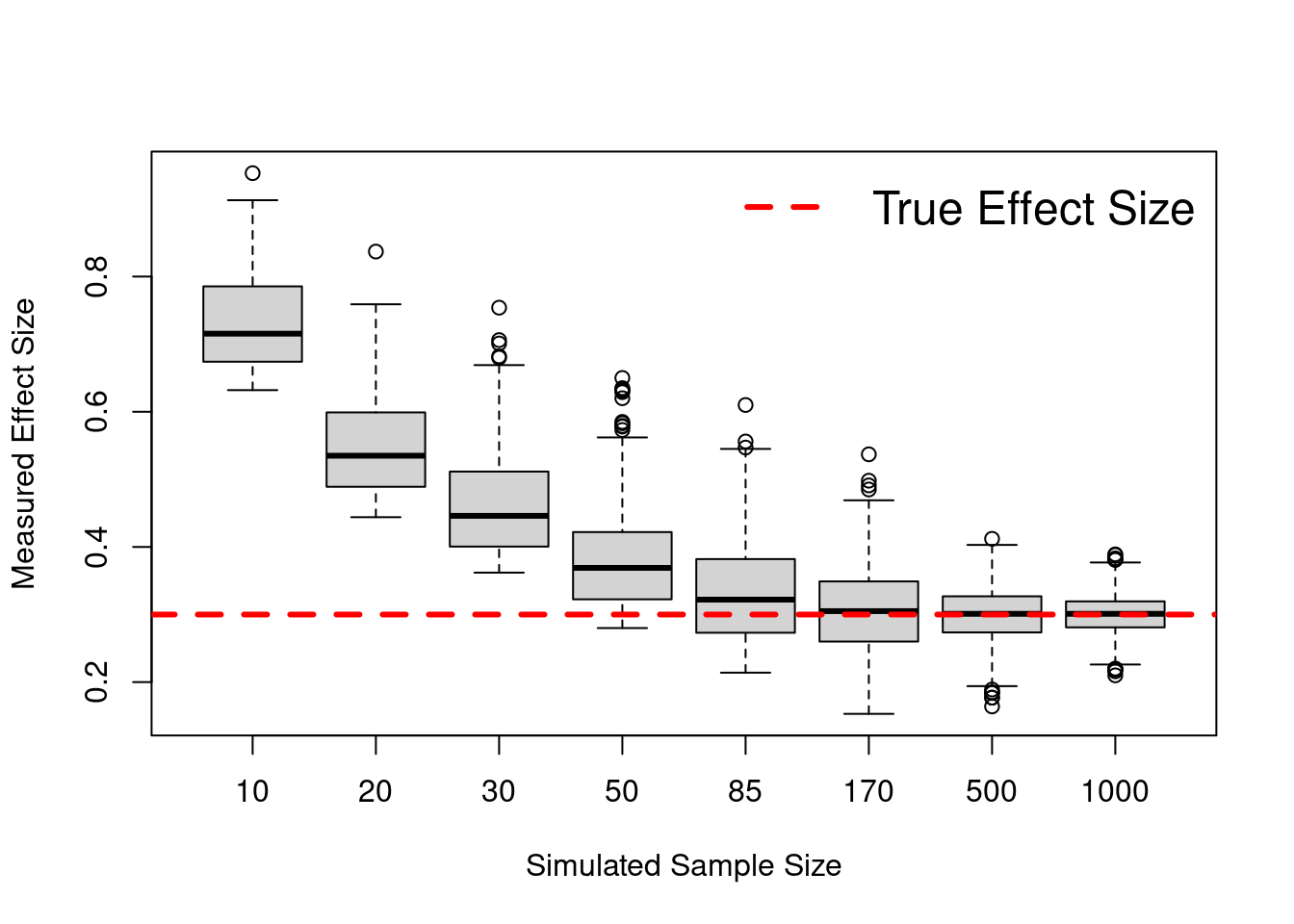
13.2 Power analysis
Usamos el paquete pwr: https://cran.r-project.org/web/packages/pwr/vignettes/pwr-vignette.html. En este tenemos un conjunto de funciones que nos permiten hacer power analysis para distintos tipos de diseños:
- pwr.p.test: one-sample proportion test
- pwr.2p.test: two-sample proportion test
- pwr.2p2n.test: two-sample proportion test (unequal sample sizes)
- vpwr.t.test: two-sample, one-sample and paired t-tests
- pwr.t2n.test: two-sample t-tests (unequal sample sizes)
- pwr.anova.test: one-way balanced ANOVA
- pwr.r.test: correlation test
- pwr.chisq.test: chi-squared test (goodness of fit and association)
- pwr.f2.test: test for the general linear model
En cada uno de ellos podemos modificar varios parámetros. El parámetro que dejemos sin especificar será el que se calculará:
- k = groups
- n = participants (in each group)
- f = effect size
- sig.level = sig level
- power = power
- etc.
Conventional effect sizes for all tests in the pwr package (SOURCE: https://cran.r-project.org/web/packages/pwr/vignettes/pwr-vignette.html):
| Test | small | medium | large |
|---|---|---|---|
| tests for proportions (p) | 0.2 | 0.5 | 0.8 |
| tests for means (t) | 0.2 | 0.5 | 0.8 |
| chi-square tests (chisq) | 0.1 | 0.3 | 0.5 |
| correlation test (r) | 0.1 | 0.3 | 0.5 |
| anova (anov) | 0.1 | 0.25 | 0.4 |
| general linear model (f2) | 0.02 | 0.15 | 0.35 |
Libraries
if (!require('pwr')) install.packages('pwr'); library('pwr')Code
#T-test
pwr.t.test(n = , d = 0.8, sig.level = 0.05, power = 0.95, type = c("two.sample"))##
## Two-sample t test power calculation
##
## n = 41.59413
## d = 0.8
## sig.level = 0.05
## power = 0.95
## alternative = two.sided
##
## NOTE: n is number in *each* group#T-test
pwr.t2n.test(n1 = 150, n2 = 75, d = , sig.level = 0.05, power = 0.8)##
## t test power calculation
##
## n1 = 150
## n2 = 75
## d = 0.3979226
## sig.level = 0.05
## power = 0.8
## alternative = two.sided#Anova
pwr.anova.test(k = 3, n = , f = .25, sig.level = 0.05, power = .8)##
## Balanced one-way analysis of variance power calculation
##
## k = 3
## n = 52.3966
## f = 0.25
## sig.level = 0.05
## power = 0.8
##
## NOTE: n is number in each group#Effect size that we can detect
pwr.anova.test(k = 6, n = 42, sig.level = 0.05, power = .95)##
## Balanced one-way analysis of variance power calculation
##
## k = 6
## n = 42
## f = 0.283334
## sig.level = 0.05
## power = 0.95
##
## NOTE: n is number in each group#Sample size needed to detect an effect of f=0.5
pwr.anova.test(k = 2, f = 0.5, sig.level = 0.05, power = .95)##
## Balanced one-way analysis of variance power calculation
##
## k = 2
## n = 26.9892
## f = 0.5
## sig.level = 0.05
## power = 0.95
##
## NOTE: n is number in each group# General linear model
# See example in https://cran.r-project.org/web/packages/pwr/vignettes/pwr-vignette.html
pwr.f2.test(u = 2, f2 = 0.3/(1 - 0.3), sig.level = 0.001, power = 0.8)##
## Multiple regression power calculation
##
## u = 2
## v = 49.88971
## f2 = 0.4285714
## sig.level = 0.001
## power = 0.8 # n=v+u+1. Therefore he needs 50 + 2 + 1 = 53 student records.13.2.1 Longitudinal designs
Calculate sample size for longitudinal designs
Libraries
if (!require('powerMediation')) install.packages('powerMediation'); library('powerMediation')Code
ssLong(es = 0.5, # effect size of the difference of mean change.
rho = 0.5, # correlation coefficient between baseline and follow-up values within a treatment group.
alpha = 0.05,
power = 0.95)## [1] 10413.3 Anonimizar participantes - Encriptar IDs
Con safer podemos encriptar cualquier cosa.
Con hashids solo podemos encriptar IDs numericos.
Libraries
if (!require('hashids')) install.packages('hashids'); library('hashids')
if (!require('safer')) install.packages('safer'); library('safer')
if (!require('dplyr')) install.packages('dplyr'); library('dplyr')13.3.1 Encriptando y desencriptando un elemento
# No acepta acentos, ñs, etc.
h = hashid_settings(salt = 'this is my salt')
# Numbers to encode
Plain_ID = c(12345678); Plain_ID## [1] 12345678a = encode(Plain_ID, h); a #"BZ9RNV"## [1] "BZ9RNV"decode("BZ9RNV", h) #12345678## [1] 1234567813.3.2 Encriptando y desencriptando la columna RUT de una DB completa.
Generamos aleatoriamente numeros de identificacion “similares” a RUTs, los encriptamos, y despues desencriptamos.
# Cargamos librerias y leemos DB
if (!require('hashids')) install.packages('hashids'); library('hashids')
if (!require('dplyr')) install.packages('dplyr'); library('dplyr')
# Pseudo RUTs generadas aleatoriamente
data = round(runif(100)*10000000) %>% as_tibble() %>%
dplyr::rename(RUT = value)BUG: When building book with Bookdown, it ignores the rowwise() argument!?
h = hashid_settings(salt = 'this is my salt')
# Encriptamos
data = data %>%
dplyr::rowwise() %>%
mutate(RUT_Encripted = encode(RUT, h))
# Desencriptamos
data = data %>%
dplyr::rowwise() %>%
mutate(RUT_Decripted = decode(RUT_Encripted, h))
# Mostramos los resultados
data %>% select(RUT_Encripted, RUT_Decripted, RUT)## # A tibble: 100 x 3
## # Rowwise:
## RUT_Encripted RUT_Decripted RUT
## <chr> <dbl> <dbl>
## 1 8RExa 2832261 2832261
## 2 l9aMNQ 7583306 7583306
## 3 WqOm93 3955537 3955537
## 4 xKXQoL 8587857 8587857
## 5 aporP 2879598 2879598
## 6 EOL3By 9179372 9179372
## 7 6g79nq 5208890 5208890
## 8 LK2er 1422724 1422724
## 9 jr47mN 5975451 5975451
## 10 BQ1jBQ 9688978 9688978
## # … with 90 more rows13.3.3 Encriptando algo que no sean numeros
if (!require('safer')) install.packages('safer'); library('safer')
if (!require('dplyr')) install.packages('dplyr'); library('dplyr')
temp <- encrypt_string("hello, how are you", key = "mi clave secreta")
decrypt_string(temp, "mi clave secreta")
KEY = "my super secret key"
# Pseudo RUTs generadas aleatoriamente
data = round(runif(100)*10000000) %>% as_tibble() %>%
dplyr::rename(RUT = value)
# **BUG: When building book with Bookdown, it ignores the rowwise() argument!?**. Sin este argumento falla la funcion
data %>%
mutate(RUT = paste0("x", RUT)) %>%
# Encrypt
rowwise() %>%
mutate(ID = encrypt_string(as.character(RUT), key = KEY)) %>%
select(RUT, ID) %>%
# Decrypt
mutate(ID2 = decrypt_string(ID, key = KEY)) 13.4 Match participants
Usaremos la siguiente Base de datos
# Cargamos librerias y leemos DBif (!require('pacman')) install.packages('pacman'); library('pacman')
if (!require('optmatch')) install.packages('optmatch'); library('optmatch')
if (!require('readr')) install.packages('readr'); library('readr')
if (!require('dplyr')) install.packages('dplyr'); library('dplyr')
# Archivo de data crudos
# RUTs y Edades generadas aleatoriamente
data = read_csv("Data/11-Experimental_design/Match_participants.csv")
head(data)## # A tibble: 6 x 7
## ID RUT Sexo Edad Ocupacion Educacion Porcentaje_ahor…
## <dbl> <dbl> <chr> <dbl> <chr> <chr> <dbl>
## 1 1 588006 Mujer 29 ASISTENTE DE TESORERIA Técnica 10
## 2 2 188576 Mujer 21 Ingeniero Comercial Maestría 50
## 3 3 206609 Hombre 46 trabajador Técnica 5
## 4 4 404898 Mujer 52 Psicóloga Maestría 0
## 5 5 533281 Mujer 63 Controller Postgrados Ej… Maestría 4
## 6 6 60824 Hombre 29 Ingeniro Comercial Maestría 8Preparamos la base de datos
# Umbrales para ahorradores y no ahorradores
Criterio_ahorradores = 20
Criterio_no_ahorradores = 2
# Mostramos histograma
hist(data$Porcentaje_ahorro, breaks = 100)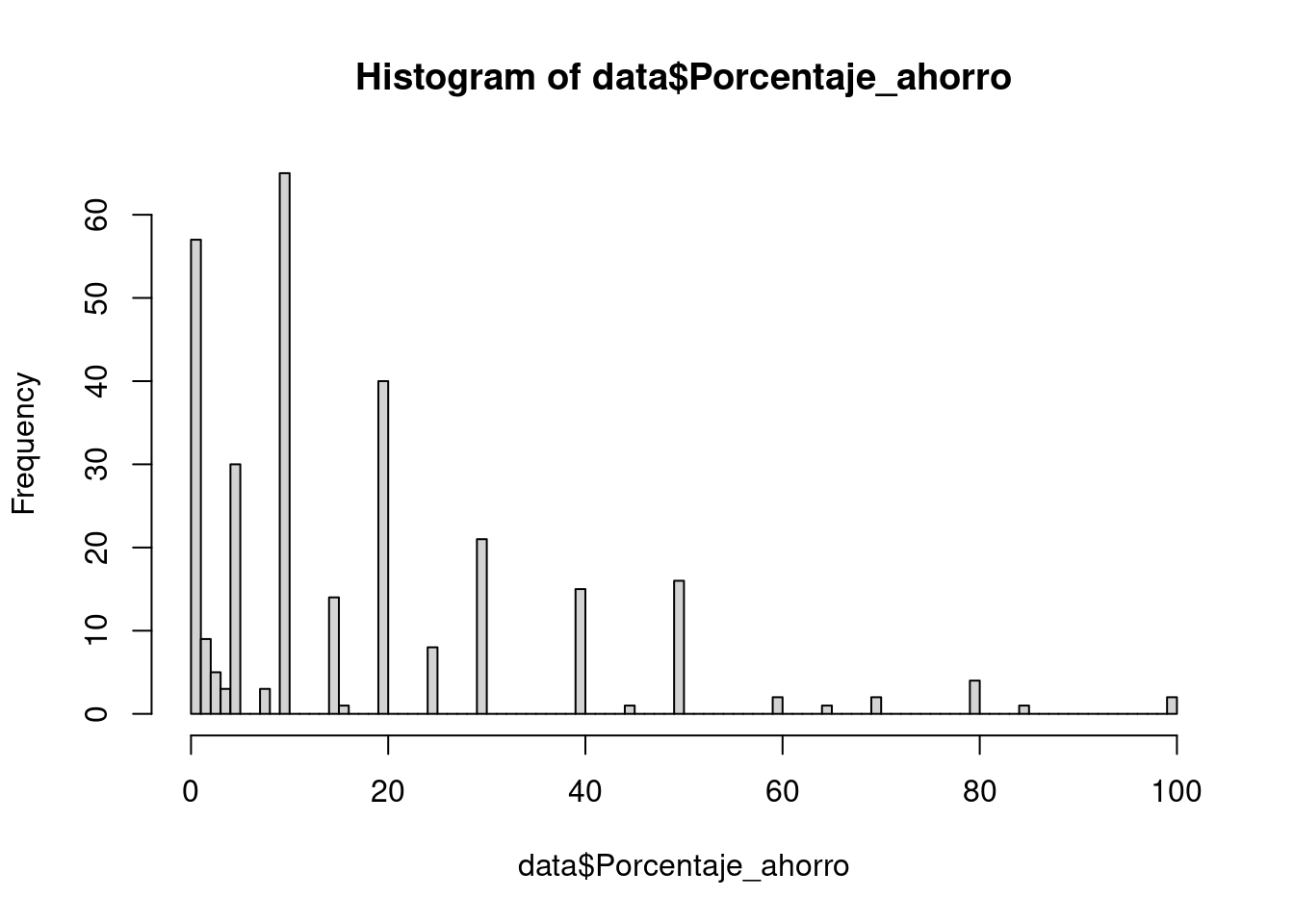
# Creamos grupos de Ahorradores (0) y No ahorradores (1)
data$Ahorro = ifelse(data$Porcentaje_ahorro <= Criterio_no_ahorradores, 1,
ifelse(data$Porcentaje_ahorro >= Criterio_ahorradores, 0, NA
))
# Cuantos tenemos en cada grupo?
table(data$Ahorro)##
## 0 1
## 113 66#Limpiamos NAs en Ahorro y RUTs duplicados
data = data[complete.cases(data),]
data = data[!duplicated(data$ID),]
#Eliminamos a los Estudiantes
data = data[!grepl("studiante", data$Ocupacion),]
#Recreamos columna ID para los registros que quedan
data$ID = seq(1:nrow(data))Realizamos el matching
TODO: Como evaluamos si el matching es bueno?
#Creamos base de parejas
parejas = as.data.frame((optmatch::pairmatch(Ahorro ~ Edad + Sexo + Educacion, data = data)))
# caliper establece limites para los matchings
# print(pairmatch(Ahorro ~ Edad + Sexo + Educacion, caliper = 4.9, data = data), grouped = TRUE)
#Como evaluamos si es buena la seleccion?
# summary(lm(Ahorro ~ Edad + Sexo + Educacion, data = data))
#Añadimos columna ID y renombramos columnas
parejas$ID = seq(1:nrow(data))
colnames(parejas) = c("Group", "ID")
#Combinamos ambas bases y limpiamos
datos_combinados = merge(data, parejas, by = "ID")
datos_combinados = datos_combinados[complete.cases(datos_combinados),]
#Ordenamos por Num de grupo y Ahorro
datos_finales = datos_combinados %>% arrange(Group, Ahorro)#Cuantos en cada grupo?
table(datos_finales$Ahorro)##
## 0 1
## 58 58# datos_finales %>% dplyr::select(Edad, Sexo, Educacion, Porcentaje_ahorro)13.5 Determine distribution
Which ditribution best fits our data.
if(!require('fitdistrplus')) install.packages('fitdistrplus'); library('fitdistrplus')
if(!require('logspline')) install.packages('logspline'); library('logspline')x <- c(37.50,46.79,48.30,46.04,43.40,39.25,38.49,49.51,40.38,36.98,40.00,
38.49,37.74,47.92,44.53,44.91,44.91,40.00,41.51,47.92,36.98,43.40,
42.26,41.89,38.87,43.02,39.25,40.38,42.64,36.98,44.15,44.91,43.40,
49.81,38.87,40.00,52.45,53.13,47.92,52.45,44.91,29.54,27.13,35.60,
45.34,43.37,54.15,42.77,42.88,44.26,27.14,39.31,24.80,16.62,30.30,
36.39,28.60,28.53,35.84,31.10,34.55,52.65,48.81,43.42,52.49,38.00,
38.65,34.54,37.70,38.11,43.05,29.95,32.48,24.63,35.33,41.34)
descdist(x, discrete = FALSE)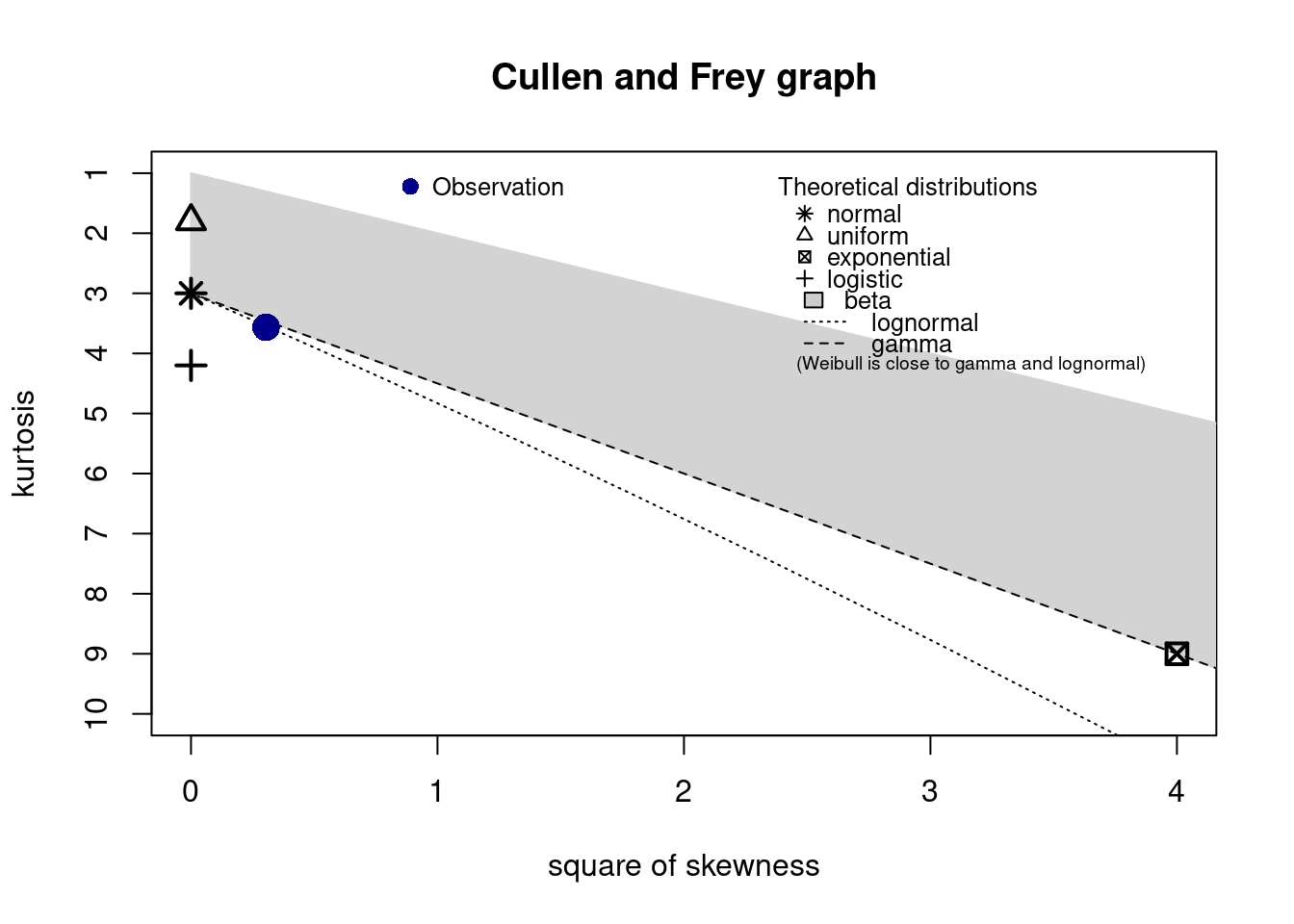
## summary statistics
## ------
## min: 16.62 max: 54.15
## median: 40.38
## mean: 40.28434
## estimated sd: 7.420034
## estimated skewness: -0.551717
## estimated kurtosis: 3.565162